Hi,
I am new to DeepChem community.
I have been trying to run the revised code from the web tutorial (pdbbind_rf.py). However, I could not load and featurize the pdbbind data from Molnet for some reason.
It seems that other people asked about the similar issue some time ago, but I cannot find the solution to this issue.
Please, refer to the line of code (from the tutorial) and the portion of the error message shown in the snapshot.
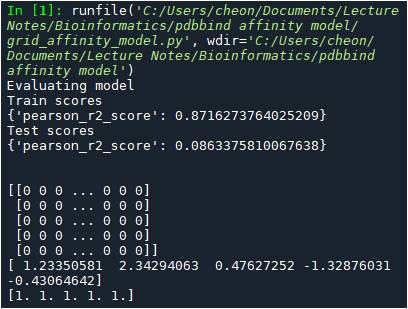
tasks, datasets, transformers = dc.molnet.load_pdbbind(featurizer=grid, splitter=“random”, subset=“core”)
Upon execution, I get the 193 lines of the following warning message:
WARNING: deepchem.feat.base_classes: Failed to featurize datapoint 0. Appending empty array.
WARNING: deepchem.feat.base_classes: Failed to featurize datapoint 1. Appending empty array.
…
WARNING: deepchem.feat.base_classes: Failed to featurize datapoint 192. Appending empty array.
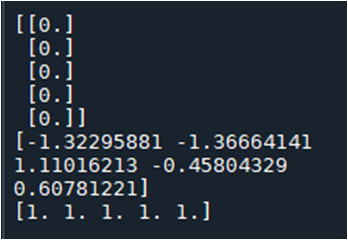
When printing out the 5 data from each dataset, I get the following:
print(train_dataset.X[0:5])
print(train_dataset.y[0:5])
print(train_dataset.w[0:5])
It would be great if someone can provide me with a solution to this challenge.
Thank you in advance.