I was selected as the contributor for the project “Torch Compile and PyTorch 2.2.0” for Google Summer of Code 2024 under DeepChem. The goal of this project — mentored by Arun Pa and Dr. Bharath Ramsundar — was to integrate the torch.compile() into the DeepChem library. By leveraging this model optimization feature, DeepChem users can now benefit from significantly improved performance during both the training and inference stages of their deep learning models.
PyTorch introduced the torch.compile() function in PyTorch 2.0 to allow faster training and inference of the models. The function works by compiling PyTorch code into optimised kernels using a JIT(Just in Time) compiler. Different models show varying levels of improvement in run times depending on their architecture and batch size when compiled. Compared to existing methods like TorchScript or FX tracing, compile() also offers advantages such as the ability to handle arbitrary Python code and conditional graph-breaking flow of the inputs to the models. This allows compile() to work with minimal or no code modification to the model. For more information on the compilation process, refer to PyTorch2.0 Introductory Blog — which does a deep dive into the compilation process, technical decisions and future features for the compile function.
As part of this project, I integrated torch.compile() into Deepchem’s TorchModel class allowing all PyTorch models in DeepChem to be compiled. Additionally, the tutorial “Compiling Deepchem Torch Models” was also added to DeepChem showing how to compile the models and benchmark them. The weekly progress on the project was tracked on this forum thread. You can find the official GSoC project page of this project here.
Connect With Me:
Twitter: https://x.com/gaushn_
GitHub: https://github.com/gauthamk02
Website: https://gauthamk02.github.io/
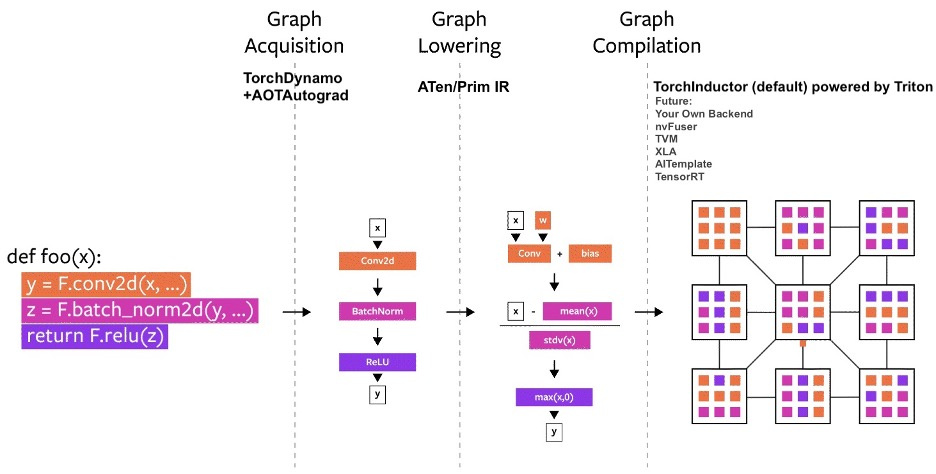
Overview of the compilation Process. Image taken from PyTorch2.0 Introductory Blog
Pull Requests
Adding support for torch.compile() and its tutorial to DeepChem involved several PRs which are:
- Initial implementation of the compile function and corresponding tests - link
This PR addedcompile()function to theTorchModelclass and also added the corresponding unit tests. The implementation of the function in this PR only allowedinductorbackend anddefault,max-autotune-no-cudagraphsoptions formodeparameter to be used as these options don’t need any additional dependencies. - Adding remaining backends and modes to the function and simplifiying it - link
All the restrictions on the backends and modes on the function was removed in this PR. The users will get an error message if a configuration is used that requires additional dependencies. - Add the tutorial for compiling Deepchem models - link
The tutorial for compiling models was added to the DeepChem Tutorials. The tutorial covers:- Steps to compile DeepChem models.
- The basic theory behind model compilation.
- Methods for comparing the performance of compiled and uncompiled models for training and inference.
- Key considerations for optimizing model compilation.
- Modify the tutorial to include more contextual information - link
The tutorial was modified to include more contextual information about the need of optimization.
Benchmarking Performance Improvements
The below results were taken from the tutorial, “Compiling Deepchem Torch Models”, which was added as part of this GSoC project to the Deepchem Tutorials. Refer to the tutorial for the complete code for benchmarking, training and obtaining these results.
To check the performance improvement after compilation, DMPNN model along with Freesolv dataset was used in the tutorial. The uncompiled execution is called Eager Mode and the compiled execution is called Compiled Mode in following results.
Kernel compilations in compiled models cause a significant performance overhead in the starting few steps. To account for this, median values are used as the performance metric when calculating speedup. After the kernels are fully compiled, the time taken by the compiled model per batch stabilises below the uncompiled model. This is because the compilation is done JIT when the model is first run and the optimized kernels are generated after a few passes.
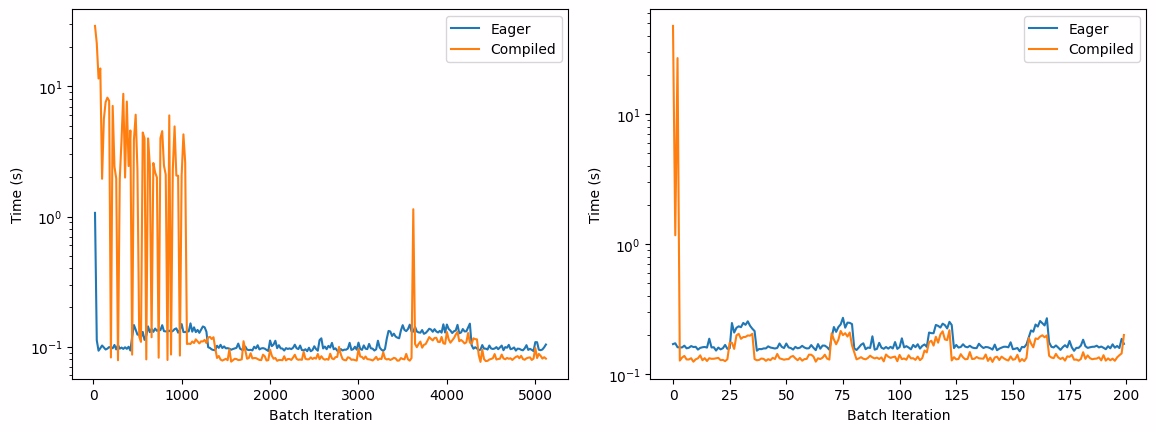
Performance comparison of average Eager and Compiled execution time of DMPNN model over 20 steps for training(left) and inference(right). Note the initial overhead in Compiled mode due to JIT compilation, followed by consistent performance gains in subsequent steps.
Results:
| Total Time | Median Time Per Batch |
Median Speedup | |||
|---|---|---|---|---|---|
| Eager | Compiled | Eager | Compiled | ||
| Training | 29.18 s | 243.32 s | 0.100 s | 0.084 s | 18.69% |
| Inference | 35.30 s | 104.21 s | 0.161 s | 0.133 s | 21.38% |
The benchmarking results show a clear speedup in the median time per batch for both training and inference after model compilation, demonstrating the effectiveness of torch.compile() in optimizing performance.
Other Works
During the rest of the program, I have worked on other tasks in DeepChem such as profiling the fit() function and solving other open issues. The profiling was done using cProfile and visualized using SnakeViz. However, I am not including the details of that in this report as it was not part of the original GSoC project. You can refer to this document for the results, inferences and code for profiling.
