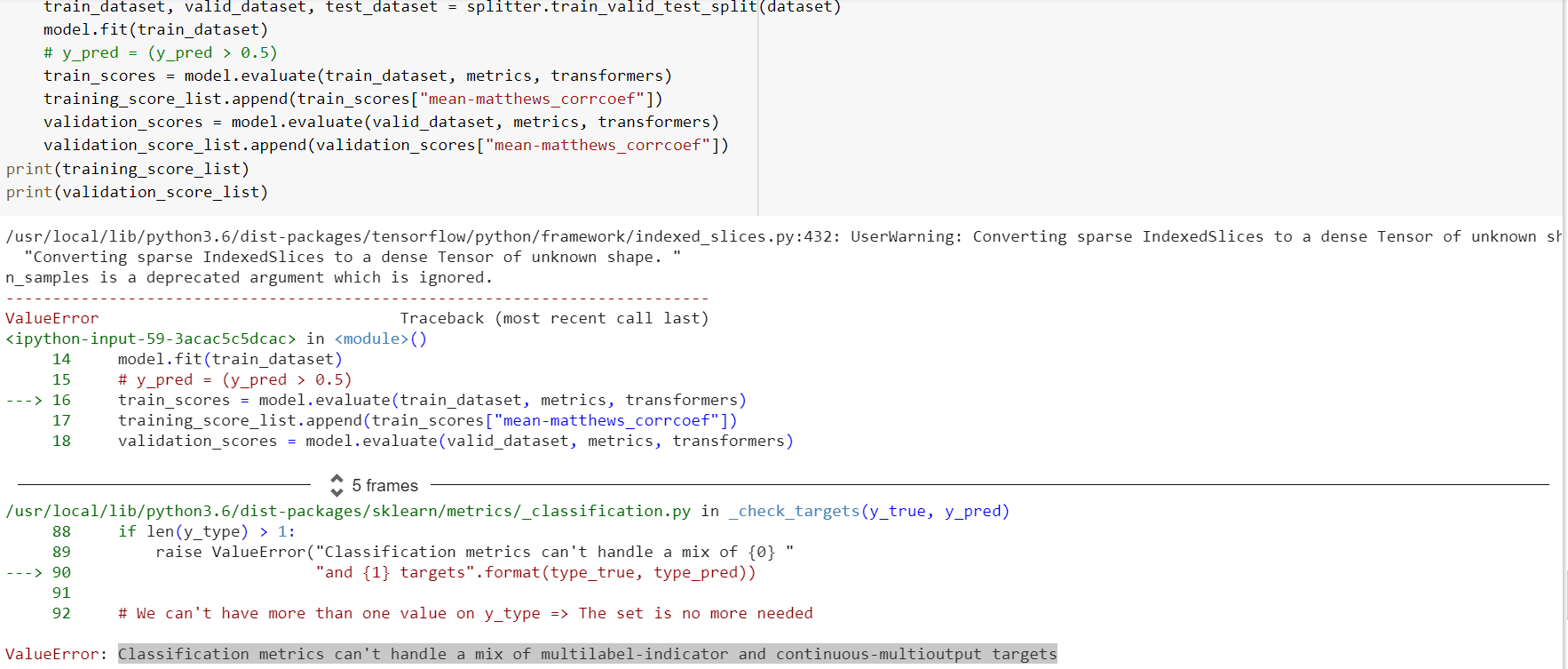
Hi there, I am trying to evaluate the GraphConv Model using
metric = dc.metrics.Metric(dc.metrics.roc_auc_score, np.mean, mode=“classification”)
train_scores = model.evaluate(train_dataset, [metric])
but am getting an “IndexError: index 123 is out of bounds for axis 1 with size 2”.
I copied the code below. Does anybody have an idea how to fix that?
Error:
File “/Users/ingrid/.conda/envs/ml-compound-clustering/lib/python3.7/site-packages/deepchem/metrics/ init .py”, line 30, in to_one_hot
y_hot[np.arange(n_samples), y.astype(np.int64)] = 1
IndexError: index 123 is out of bounds for axis 1 with size 2
import numpy as np
import matplotlib.pyplot as plt
import deepchem as dc
from rdkit import Chem
from rdkit.Chem import Draw
import pandas as pd
from itertools import islice
from IPython.display import Image, display
from sklearn.model_selection import train_test_split
from deepchem.models import GraphConvModel
from deepchem.utils.save import load_from_disk
Load dataset
dataset_file = ‘test.csv’
Featurize
featurizer = dc.feat.ConvMolFeaturizer()
Convert data into machine learning suitable data object
loader = dc.data.CSVLoader(tasks=[“task1”], smiles_field=“smiles”, featurizer=featurizer)
dataset = loader.featurize(dataset_file)
Initialize transformer
#transformer = dc.trans.NormalizationTransformer(transform_w=True, dataset=dataset)
#dataset = transformer.transform(dataset)
Define train, validation and test data sets
splitter = dc.splits.ScaffoldSplitter(‘test.csv’)
train_dataset, valid_dataset, test_dataset = splitter.train_valid_test_split(dataset)
model = GraphConvModel(n_tasks=1, mode=‘classification’, batch_size=50, n_classes=125)
model.fit(train_dataset, nb_epoch=100)
metric = dc.metrics.Metric(dc.metrics.roc_auc_score, np.mean, mode=“classification”)
train_scores = model.evaluate(train_dataset, [metric])