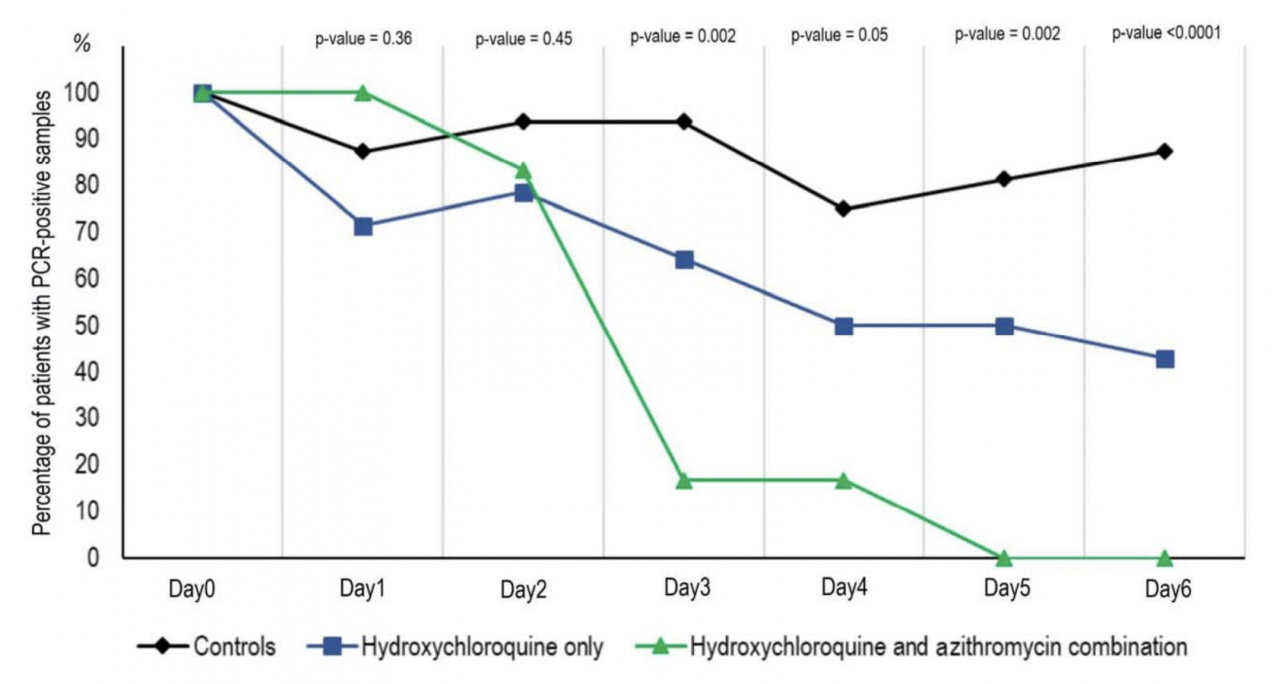
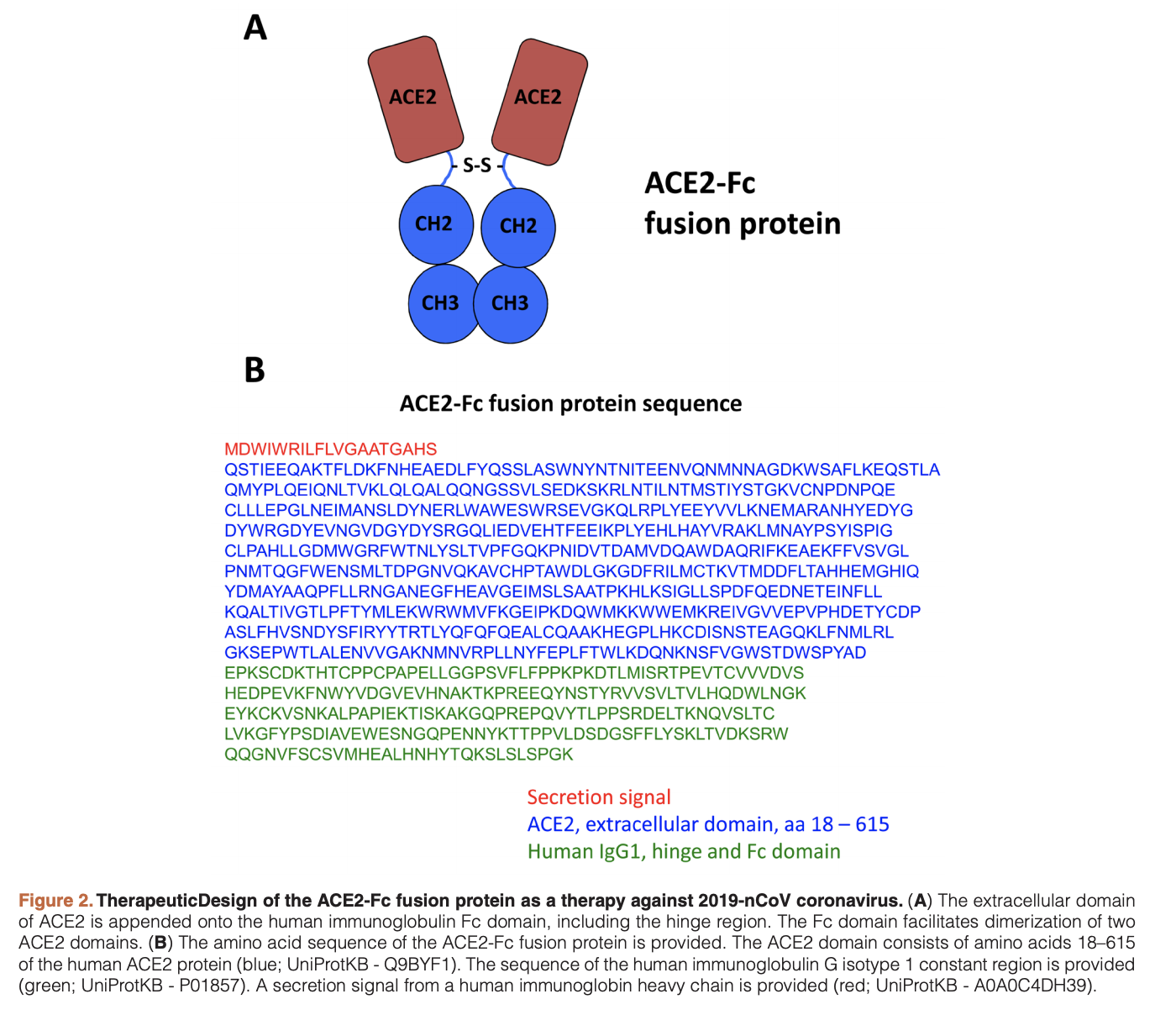
SARS-CoV-2 vaccines: status report
This review article provides an overview of current therapeutics for Covid19, with a focus on vaccine development. Trials with remdesivir and protease inhibitors are currently ongoing in China. It’s not entirely clear what remdesivir’s mechanism of action is (it’s a nucleotide analog), but it may work by terminating RNA synthesis or by leading to incorporation mutagenesis. In addition a number of trials are being tried with HIV inhibitors such as lopinavir and ritonavir. Other treatment options being tried include dosing of human recombinant ACE2 and the antiviral arbidol. Human polyclonal IgG from transgenic cows was used for MERS-CoV and may also work here.
We know from SARS-CoV-1 that the spike protein is a good target for vaccine design. Several SARS-CoV-1 vaccines have been designed and tested in animal models. Vaccination with live virus resulted in lung damage in mice. Only a small number of SARS-CoV-1 vaccines made it to phase I trials before funding dired up. It’s possible that these SARS-CoV-1 vaccines might protect against SARS-CoV-2, but these aren’t currently available for use since they weren’t developed past phase I. There are some vaccines against MERS-CoV available, but these aren’t likely to protect against SARS-CoV-2.
An mRNA based vaccine (which expresses the target antigen in vivo) after injection of mRNA in lipiid nanoparticles recently started a phase I clinical trial. Other efforts in preclinical phases include recombinant protein based vaccines (focused on the spike protein), viral vector based vaccines (focused on the spike protein), DNA vaccines (focused on the spike protein), live attenuated vaccines, and inactivated virus vaccines. It’s not yet clear which of these is the best.
The paper tries to address the question of why it takes so long. Many of the technologies used for the vaccines are new and need to be tested for safety. This typically involves a test in an animal model of the virus. However, this is challenging since the virus doesn’t grow in wildtype mice and only induces mild disease in transgenic animals expressing human ACE2. In vitro neutralization assays can still be used to evaluate the serum from vaccinated animals even if the disease model isn’t accurate. Toxicity tests need to be performed (in rabbits typically). Usually this takes 3-6 months. Development of current Good Manufacturing Processes (cGMP) is also needed. All these processes need to be developed or amended for the SARS-CoV-2 vaccines. Safety and efficacy are evaluated in the usual Phase I, II, III trials.
Another challenge is that production capacity to produce sufficient vaccine might be challenging. For mature technologies such as inactivated or live attenuated vaccines, this already exists, but for novel technologies like mRNA, this may be challenging.
Finally, distributing the vaccine will take time. Vaccinating a large portion of the population may take weeks. More than one dose of the vaccine may be needed per person. This usually takes a prime-boost form, with immunity only achieved 1-2 weeks after the second. Given all these challenges, realistic time frames seem like 12-18 months.
The paper asks what longer term solutions might be. One might be to build up production capacity for respiratory viruses. More ambitious would be development of a broadly protective vaccine. This is being attempted for influenza but considerable work would be needed to develop this further.
The paper concludes by stating that in the past funds for vaccine development have only been available with looming pandemics. Given the seriousness of the current situation, it seems worth investing in better vaccine infrastructure. It also calls for developed emergency plans to be pulled out in case of future pandemics.