I spent five weeks in joining the DeepChem as a GSoC student. I want to explain what I did in five weeks.
What is JAXChem?
JAXChem is a JAX-based deep learning library for complex and versatile chemical modelings. In GSoC, my task is building this library. Please see the project details from the previous post.
What I did
As I mentioned in this roadmap, I tried to implement GCN models and make tutorials during 1st evaluation period. The reason why I chose this topic is that the GCN(GNN) is the most popular method as an example of deep learning in the area of chemistry. I think this is a good starting point for JAXChem.
During 1st evaluation period, I implemented the two pattern GCN models with Haiku which is a simple neural network library for JAX. Haiku uses the OOP style, so our codes became more friendly for many users which use PyTorch.
- Implement the pad pattern GCN model
- PRs : https://github.com/deepchem/jaxchem/pull/3, https://github.com/deepchem/jaxchem/pull/5
- This model uses adjacency matrix (shape : (V, V)) for representing node connections
- Memory efficiency is not good, so it is difficult to treat a large graphs
- Implement the sparse pattern GCN model
- PRs : https://github.com/deepchem/jaxchem/pull/7
- This model uses adjacency list (shape : (2, E)) for representing node connections
- Memory efficiency is good, so there is a possibility of treating large graph
- PyTorch Geometric adopts this style
One of the challenging point of JAXChem is to implement the sparse pattern GCN model. The pad pattern model is simpler than the sparse pattern and the blog was published like this. If you want to confirm the difference between two models more deeply, please check this repository.
Currently, I prepared for a tutorial notebook with Tox21. If you’re interested in JAXChem,
please try the following notebook in Google Colab.
Colab Tutorial : https://github.com/deepchem/jaxchem/blob/master/notebooks/gcn_property_prediction.ipynb
Performance result
I checked training performance of the pad pattern GCN model. (Ths sparse pattern GCN model has a serious performance issue, so I skipped it.) In this time, I compared training performance of the GCN model among JAXChem, Deep Graph Library(DGL) and DeepChem.
Code:
benchmark.ipynb
Environment :
-
GPU: Tesla P100 (Google Colab)
-
CUDA/cuDNN version: 10.1 / 7.65
-
DGL version: dgl-cu101 0.4.3.post2
-
DeepChem version: 2.3.0
Measuring method :
I calculated the average time per epoch was calculated for 50 epochs.
Result :
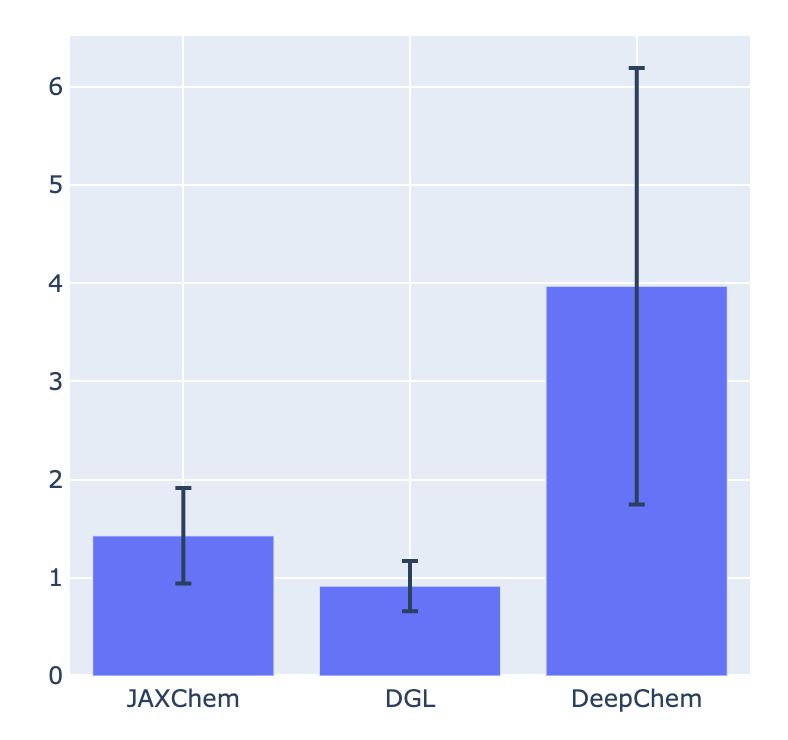
Y-axis is the average time per epoch (the unit is a second). JAXChem is not superior to DGL, but the difference is small. And then, JAXChem shows better performance than DeepChem. I think the JAXChem model still has a room to improve the performance, like rewriting the Python for-loop during training loop using JAX’s optimized functions, lax.scan or lax.fori_loop. Please stay tuned for further update!
Issues
I found the performance issue about the sparse pattern GCN model when making the Tox21 example.
The reason of the performance issue is related to this issue. The sparse pattern GCN model uses jax.ops.index_add, but a large Python “for” loop leads to a serious performance issue when using jax.ops.index_add (Training time/epoch of the Tox21 example is almost 30 times than the pad pattern.)
In order to resolve this issue, I have to rewrite training loop using lax.scan or lax.fori_loop. However, lax.scan or lax.fori_loop have some limitations like the generator/iterator doesn’t work (See this issue), so it is difficult to rewrite. Now, I’m struggling this issue and please confirm the more details in the following issue.
Performance issue : https://github.com/deepchem/jaxchem/issues/8
Next plan (during 2nd evaluation period)
According to the roadmap, I’ll be working for implementing the CGCNN model. In the next period, I will focus on supporting inorganic crystal data before Please confirm the details below.
- Build the JAXChem document (7/6 - 7/12)
- Implement a dataloader for crystal data and a new graph data class (7/13 - 7/20)
- Implement the featurizer for CGCNN (7/20 - 7/27)
- Implement the CGCNN model (7/27 - 8/3)
Finally, we welcome to contribute any chemical modelings with JAX! Please contact us if you’re interested in JAXChem.
