
I was fortunate to participate as a contributor to Google Summer of Code 2023, working with DeepChem (Open Chemistry), an Open-Source initiative that aims to democratize the use of deep learning in various fields, such as drug discovery, material science, quantum chemistry,
and biology. The project provides high-quality tools that enable researchers to use deep
learning to address complex problems in these fields.
Recently, DeepChem has been shifting its backend from TensorFlow to PyTorch. This transition requires the porting of models that were previously built in TensorFlow to PyTorch.
In this forum post, I will provide comprehensive documentation and a detailed description of all the tasks and projects I have undertaken throughout the summer period.
Project Title
Porting SeqToSeq and DTNN Model to PyTorch
This project aims to create an equivalent model for the Deep Tensor Neural Network (DTNN) [1] model and Sequence To Sequence (SeqToSeq) [2] model in PyTorch. This requires translating the TensorFlow models into their PyTorch equivalents.
Some Important Links
My GitHub: https://github.com/GreatRSingh
My LinkedIn: https://www.linkedin.com/in/rakshit-singh-ai/
DeepChem Home Page: https://deepchem.io/
DeepChem GitHub: https://github.com/deepchem/deepchem
Proposal Link: Link
Preface
Molecular property prediction plays a pivotal role in drug discovery. Recent advancements in deep learning have revolutionized this domain, offering promising alternatives that can automatically extract intricate features from molecular structures.
Deep Tensor Neural Network is based on the many-body Hamiltonian concept, which is a fundamental principle in quantum mechanics. DTNN receives a molecule’s distance matrix and membership of its atom from its Coulomb Matrix representation. Then, it iteratively refines the representation of each atom by considering its interactions with neighboring atoms. Finally, it predicts the energy of the molecule by summing up the energies of the individual atoms.
Sequence To Sequence consists of two parts called the encoder and decoder. The job of the encoder is to transform the input sequence into a single, fixed-length vector called the embedding. That vector contains all relevant information from the input sequence. The decoder then transforms the embedding vector into the output sequence. And if we keep the input and output the same then it can work as an AutoIncoder.
Contents covered in the forum
- Expected Deliverables
- DTNN Model
- Layers of DTNN Model
- Usage of DTNN Model
- SeqToSeq Model
- Layers of SeqToSeq Model
- Fixes and Additions to DeepChem
- Future Scope
- Acknowledgement
- References
Expected Deliverables
Porting Embedding, Step, and Gather Layer of DTNN Model.
Create Torch Model with utility functions of DTNN.
Porting Encoder and Decoder Layer of SeqToSeq Model.
Create Torch Model with utility functions of SeqToSeq. (In Progress)
Deep Tensor Neural Network
Encoding Molecular Representation
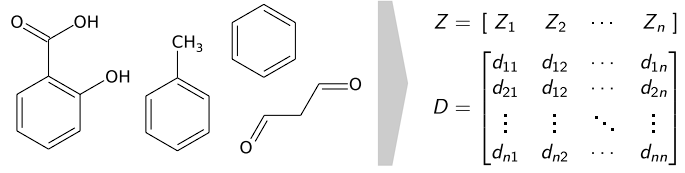
Molecules are encoded as input for the neural network by a vector of nuclear charges and an inter-atomic distance matrix. In our implementation, we use a Coulomb Matrix for this purpose. This description is complete and invariant to rotation and translation.
Network architecture.
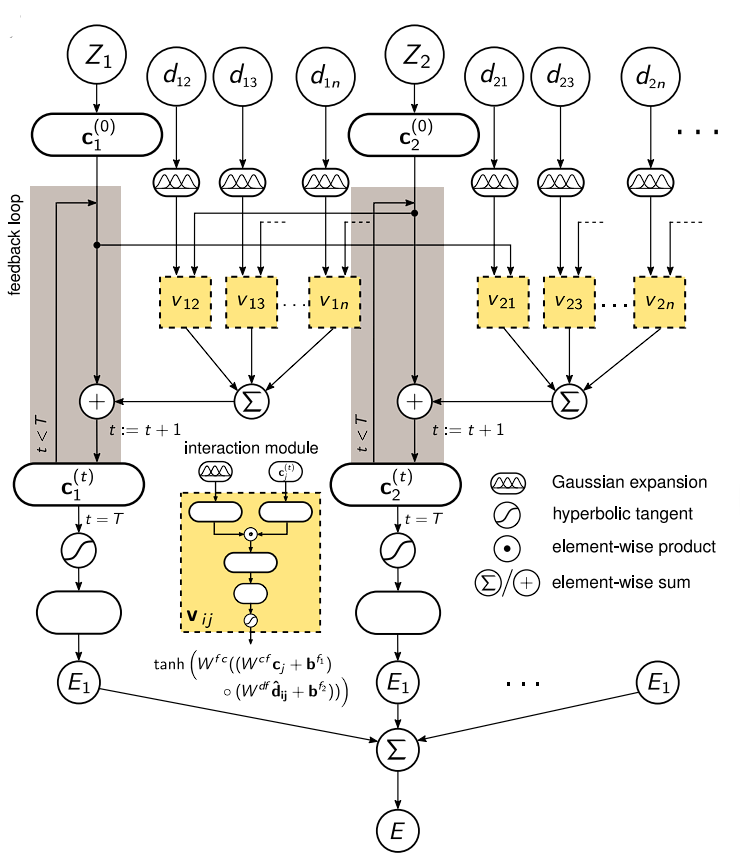
Each atom type corresponds to a vector of coefficients Ci(0) which is repeatedly refined by interactions Vij. The interactions depend on the current representation Cj(t) as well as the distance Cij to an atom j. After T iterations, an energy contribution Ei is predicted for the final coefficient vector Ci(T). The molecular energy E is the sum of all these atomic contributions.
Corresponding PRs:
- Porting DTNN Class · Pull Request #3513 +633 -7
- Porting DTNNModel Class · Pull Request #3514 +251 -5
Layers of DTNN Model
DTNNEmbedding Layer
The embedding layer creates ‘n’ number of embeddings as Initial Atomic Descriptors. According to the required weight initializer and periodic table length (Total number of unique atoms).
Corresponding PRs:
- Porting DTNNEmbedding Layer · Pull Request #3415 +108 -0
- DTNNEmbedding Layer Parameter Error Fix · Pull Request #3455 +17 -14
DTNNStep Layer
![]()
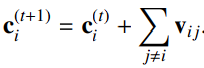
The Step Layer Encodes the atom’s interaction Vij with other atoms according to distance relationships dij. Then sum them up to get the final output Ci(t+1). Here, W are weights and b are bias.
Corresponding PRs:
- Porting DTNNStep Layer · Pull Request #3436 +201 -0
- DTNNStep Parameter error Fix · Pull Request #3454 +28 -24
DTNNGather Layer
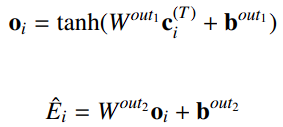
Predict Molecular Energy using atom_features and atom_membership. This Layer gathers the inputs from the step layer according to atom_membership and calculates the total Molecular Energy.
The bias parameters, as well as weights, are initially set to zero.
Corresponding PRs:
Usage of DTNN Model
The script below is an example of how to use this model:
import os
from deepchem.data import SDFLoader
from deepchem.feat import CoulombMatrix
from deepchem.models.torch_models import DTNNModel
# Load Data
dataset_file = "assets/qm9_mini.sdf'
TASKS = ["alpha", "homo"]
loader = SDFLoader(tasks=TASKS, featurizer=CoulombMatrix(29), sanitize=True)
data = loader.create_dataset(dataset_file, shard_size=100)
n_tasks = data.y.shape[1]
# Initialise the Model
model = DTNNModel(n_tasks,
n_embedding=20,
n_distance=100,
learning_rate=1.0,
mode="regression")
loss = model.fit(data, nb_epoch=250)
pred = model.predict(data)
mean_rel_error = np.mean(np.abs(1 - pred / (data.y)))
print(mean_rel_error)
Sequence To Sequence
Encoding Molecular Representation

We will be representing each word in a language as a one-hot vector, or giant vector of zeros except for a single one (at the index of the word).
Network Architecture
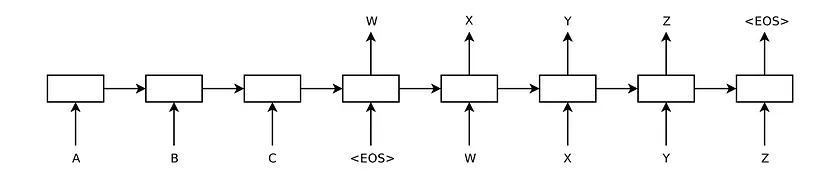
The seq2seq model consists of two subnetworks, the encoder and the decoder. At each time step, the decoder generates an element of its output sequence based on the input received and its current state, as well as updating its own state for the next time step. The input and output sequences are of fixed size.
The critical point of this model is how to get the encoder to provide the most complete and meaningful representation of its input sequence in a single output element to the decoder because this vector or state is the only information the decoder will receive from the input to generate the corresponding output.
Layers of SeqToSeq Model
EncoderRNN Layer
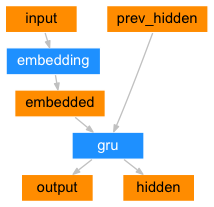
It takes input sequences and converts them into a fixed-size context vector called the “embedding”. This vector contains all relevant information from the input sequence. This context vector is then used by the decoder to generate the output sequence and can also be used as a representation of the input sequence for other Models.
Corresponding PRs:
DecoderRNN Layer
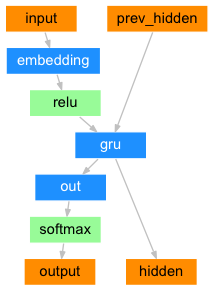
The decoder transforms the embedding vector into the output sequence. It is trained to predict the next token in the sequence given the previous tokens in the sequence. It uses the context vector from the encoder to help generate the correct token in the sequence.
Corresponding PRs:
- Porting decoder rnn ·Pull Request #3537 (Under Progress)
Fixes and Additions to DeepChem
I have made a total of 14* PRs, 86+ commits, and 5709+ additions in total.
*: Only useful PRs counted, not Work Review PR.
DeepChem suite now includes:
- A Torch Backend Based DTNN Model.
- A separate Batch Utility function.
Pull Requests to solve issues:
- Changing indent width to 4 · Pull Request #3207 +2,180 −2,073
- Adding Usage Examples to Splitters · Pull Request #3241 +127 -0
- Porting SetGather · Pull Request #3255 +192 -1
Some Works in SeqToSeq Model are remaining which will be completed in the next few weeks.
You can find my progress reports, made throughout the summer, for the project at the DeepChem Forum
Future Scope
A good upgrade to the SeqToSeq Model could be Adding an Attention Layer to it based on [3]. It has the potential to improve its performance and work on even a small amount of data.
Acknowledgment
I want to give a big shoutout to my mentors Bharath Ramsundar, Tony Davis, and Shaipranesh S, along with the amazing DeepChem community. Your guidance, suggestions, and discussions have been invaluable. Also a special mention to Aryan Amit Barsainyan and Riya Singh for always supporting me in this journey.
Being a part of this organization has provided me with real-world insights into the inner workings of ML models, especially those using graph-based neural networks. I’m committed to continuing my contributions by enhancing project implementations, addressing issues, and supporting the community as much as I can.
A special thank you to Google for making all of this possible!
References
[1] Quantum-Chemical Insights from Deep Tensor Neural Networks
[2] Sequence to Sequence Learning with Neural Networks
[3] Attention Is All You Need

