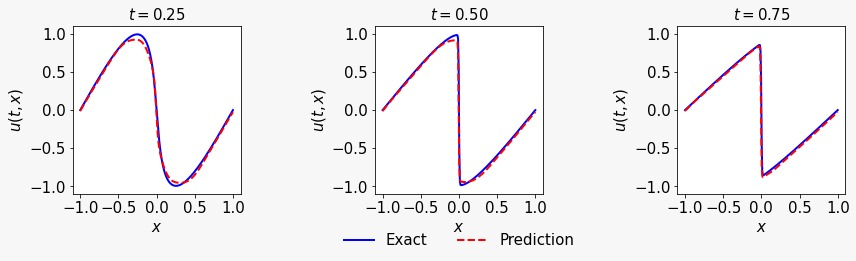
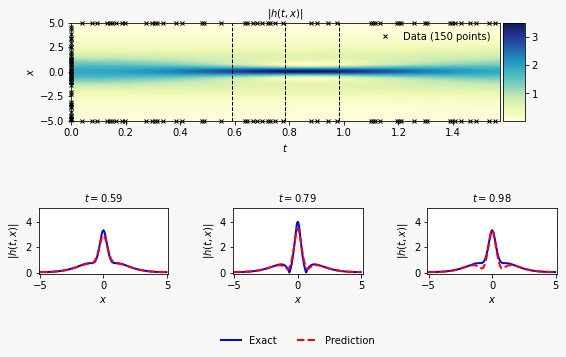
Hey everyone!
I’m Vignesh Venkataraman, an incoming final year student at IIT Roorkee, India. In the past, I have worked on building generative models for molecular strings (SMILES) using NLP and graphical machine learning with molecules.
This summer, I will be working with deepchem as a gsoc student. Some of my best learning experiences have come from contributing to deepchem in the past and I’m hoping to learn more from the program.
I will be working on integrating Jax support into the deepchem infrastructure. JAX is comparatively a new library and has received a warm welcome from the Machine Learning researchers because of core Functional approach and state management. Looking forward to learn more about Jax …