Hi,
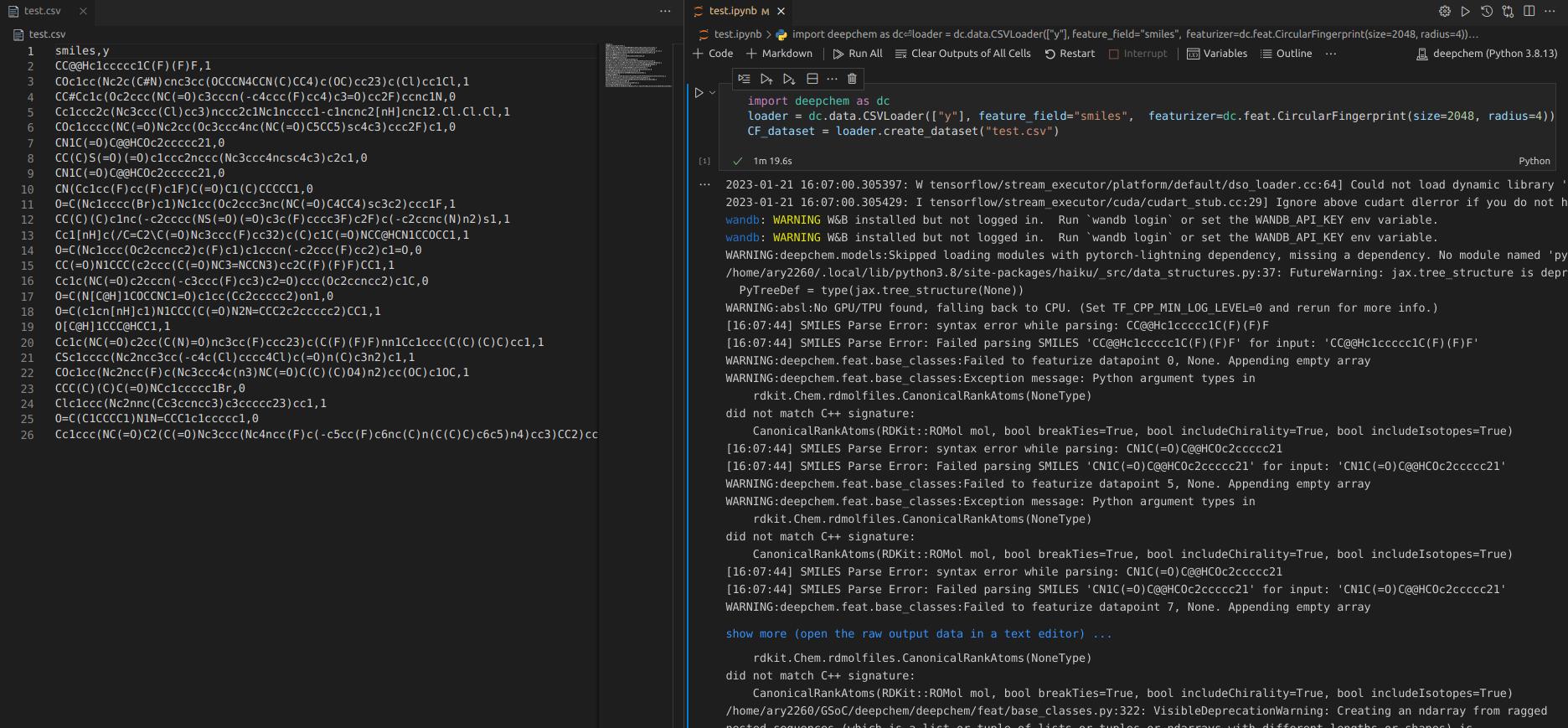
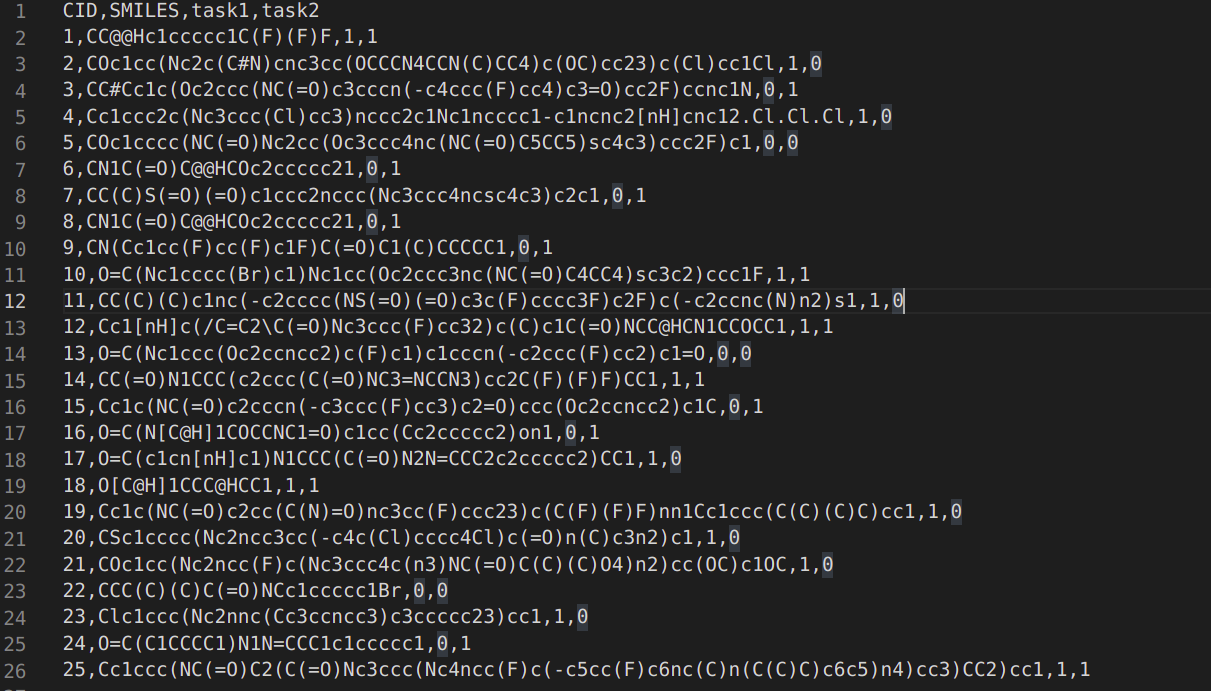
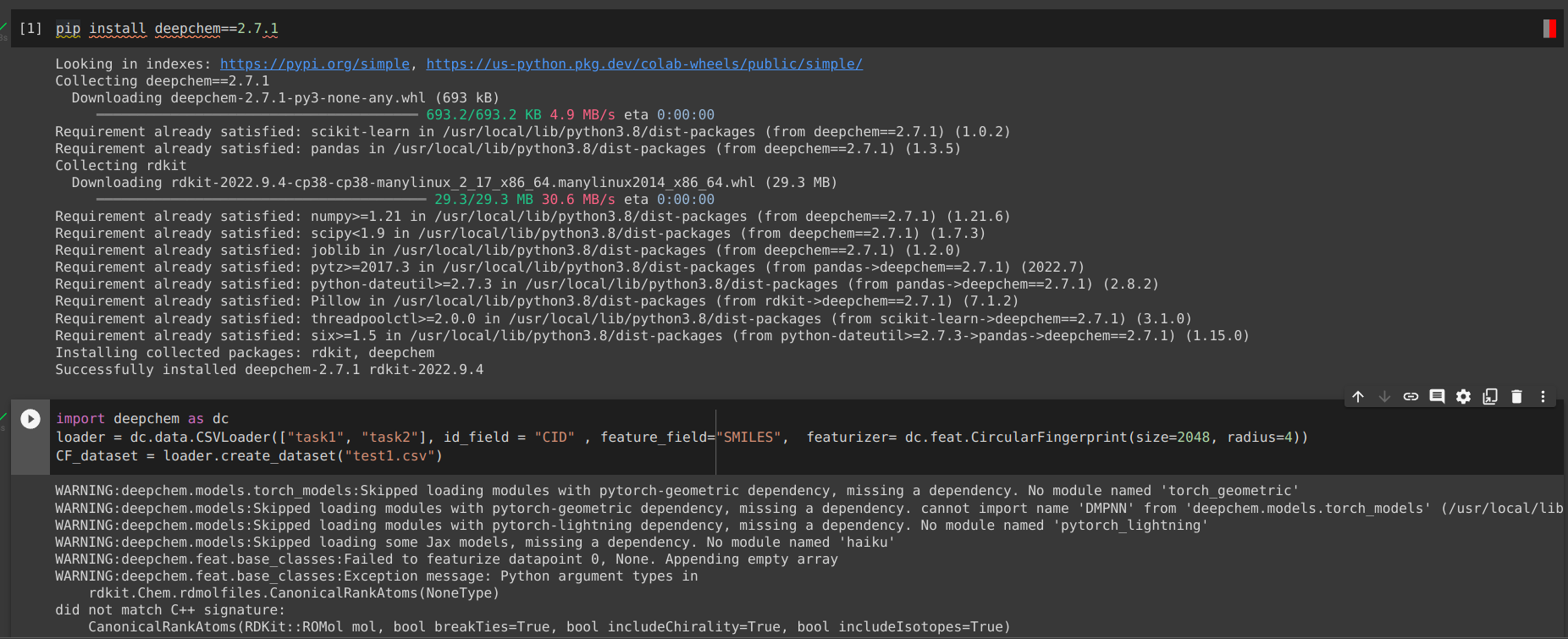
This is my first post, so please let me know if there conventions I should follow. I am featurizing a dataset created from dc.data.csvloader using the Circular Fingerprint featurizer, shown below:
import deepchem as dc
loader = dc.data.CSVLoader(["task1", "task2"], id_field = "CID" , feature_field="SMILES", featurizer= dc.feat.CircularFingerprint(size=2048, radius=4))
CF_dataset = loader.create_dataset(dataset_path)
DeepChem Version: 2.7
Issue: Featurizer failure.
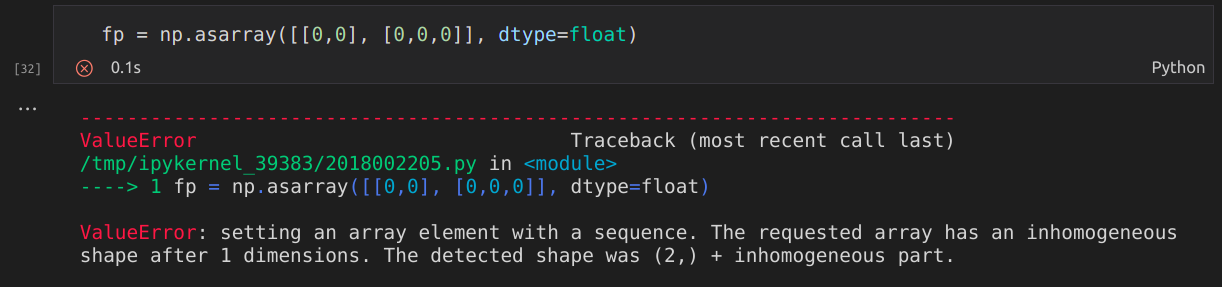
Error message: ValueError : setting an array element with a sequence. The requested array has an inhomogeneous shape after 1 dimensions. The detected shape was (8192,) + inhomogeneous part.
Please let me know if you have any suggestions. I ran this same code on DeepChem 2.5 a few months back, and had no problem. I have also tried using many other featurizers, and I get the same error except when I use RDKitDescriptors, which successfully featurizes the dataset.