Hi DeepChem community,
I run a small biotech-focused software company called Trident Bioscience and we recently put together a set of Jupyter widgets to help input, visualize, split, and analyze chemical data. We call them Trident Chemwidgets and opened up the GitHub repo earlier this week as a kind of soft launch. Chemwidgets are open-source and free to use in any capacity. I thought these tools might be of interest to this community since they can help augment and debug your DeepChem-based workflows.
You can find the Chemwidgets repository here: https://github.com/tridentbio/trident-chemwidgets
Since the package is currently pre-release, we’re looking to identify any bugs in the installation/setup/use of the package as well as any feedback from you about which features you’d like to see us build in the future. We are prioritizing features that can help our users understand and visualize their data and those that can help them debug when and why a model is training poorly. We’d love to hear any feedback you have either by email: tshimko [at] trident.bio or by raising an issue on our GitHub page. Thanks in advance for any help you can offer and I sincerely hope these widgets can be helpful to you as you build out your DeepChem models.
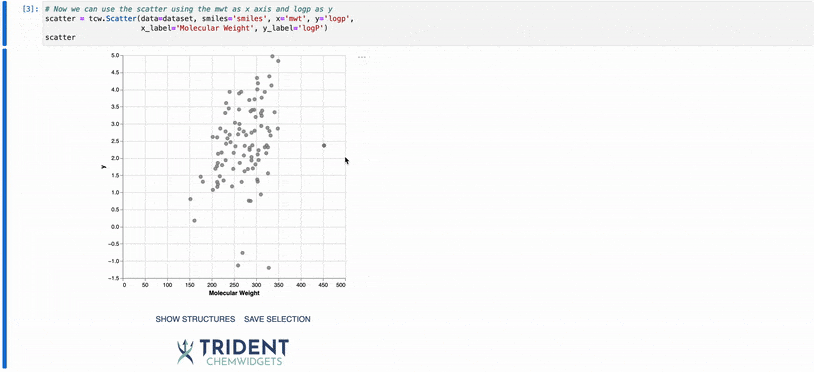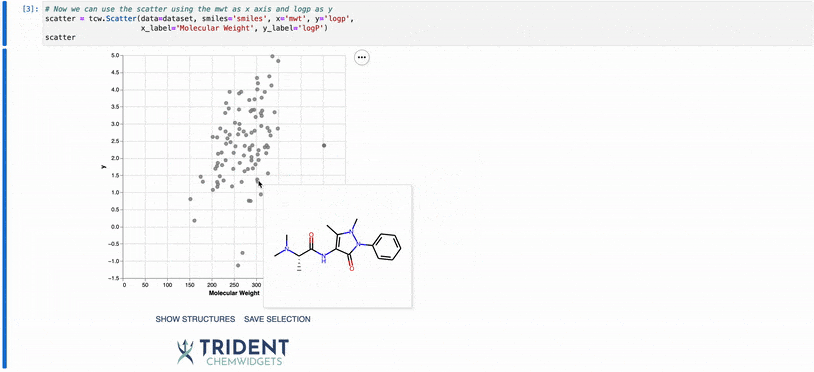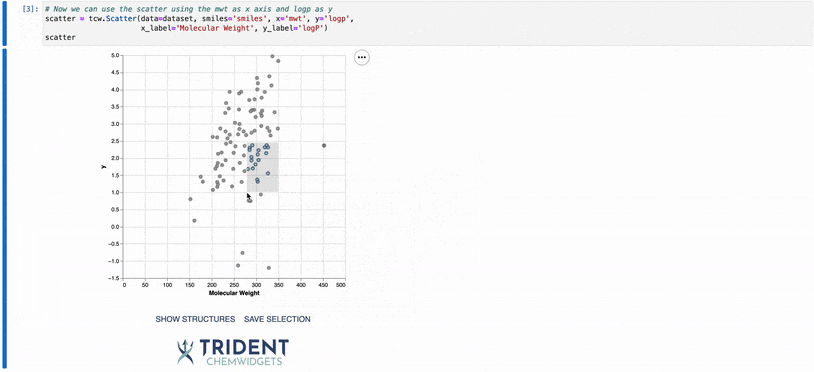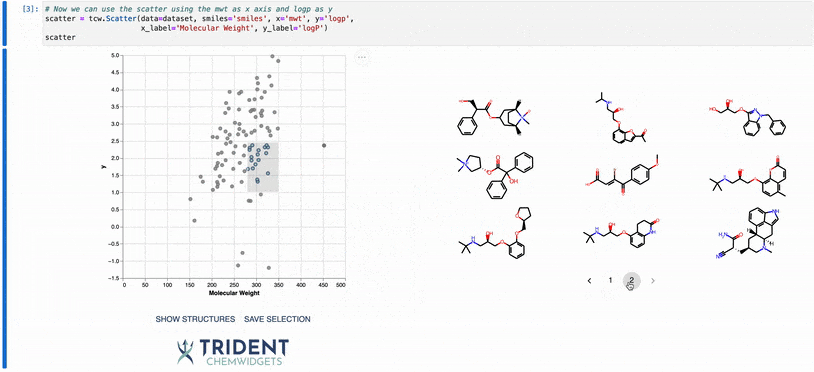
To give you a taste of what these widgets can do, I’ve included a gif of our scatter plot widget:
Thanks,
Tyler Shimko