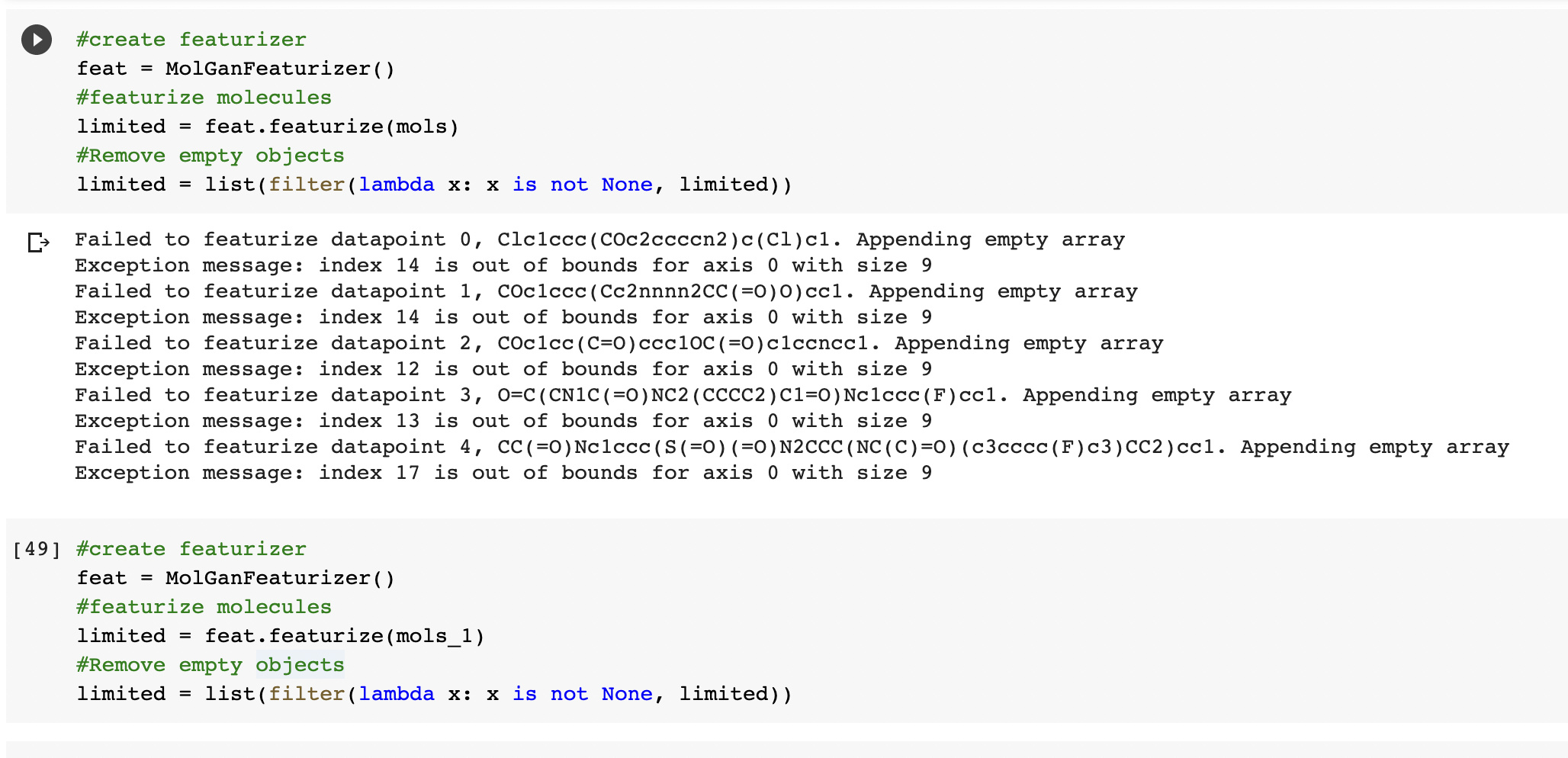
I received the same error as above even after increasing max_atom_count.
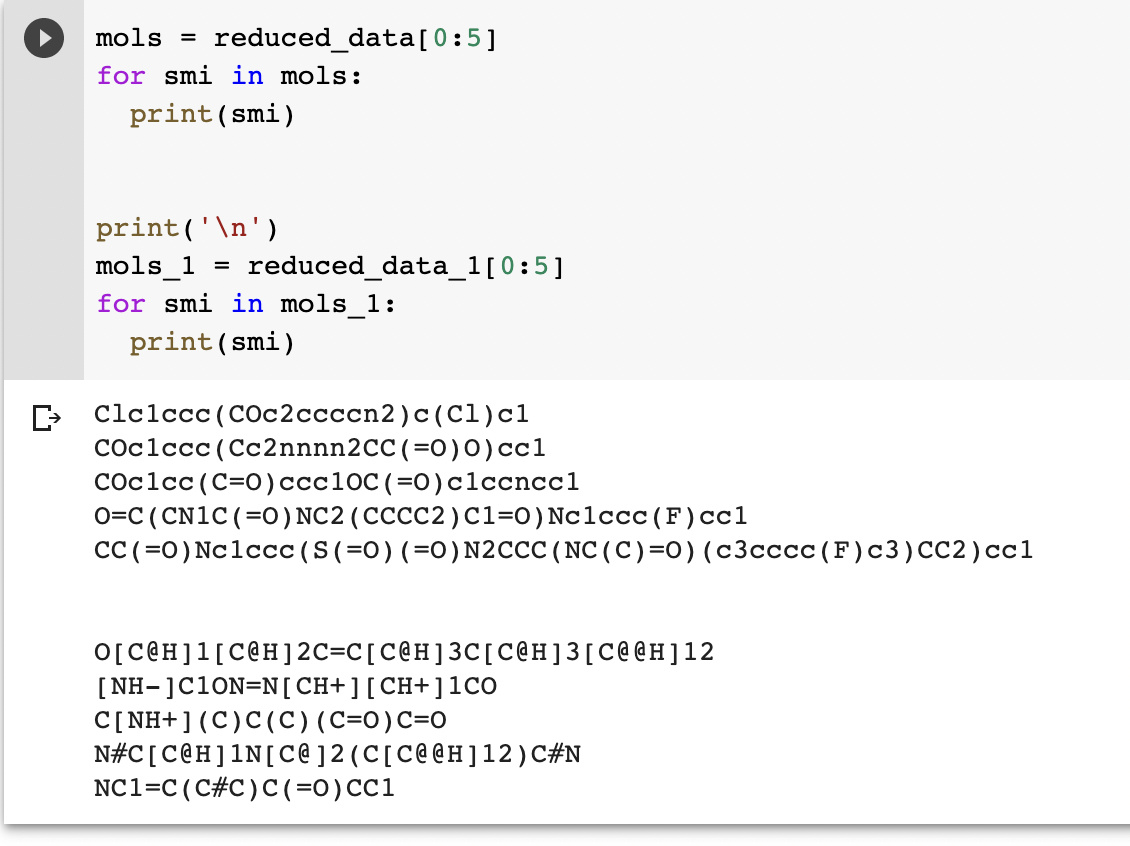
Some example SMILES I used are:
COC(=O)C1©CCCC2=C3C(=O)C(=O)C4=C(OC=C4C)C3=CC=C12
CC1COC2=C1C(=O)C(=O)C1=C3CCCC©©C3=CC=C21
Failed to featurize datapoint 3, CC(=O)c1ccccc1S(=O)(=O)c1ccccc1C(=O)O. Appending empty array
Exception message: 16
EDIT: Is there any way for me to use smiles containing S? When I removed them it worked.
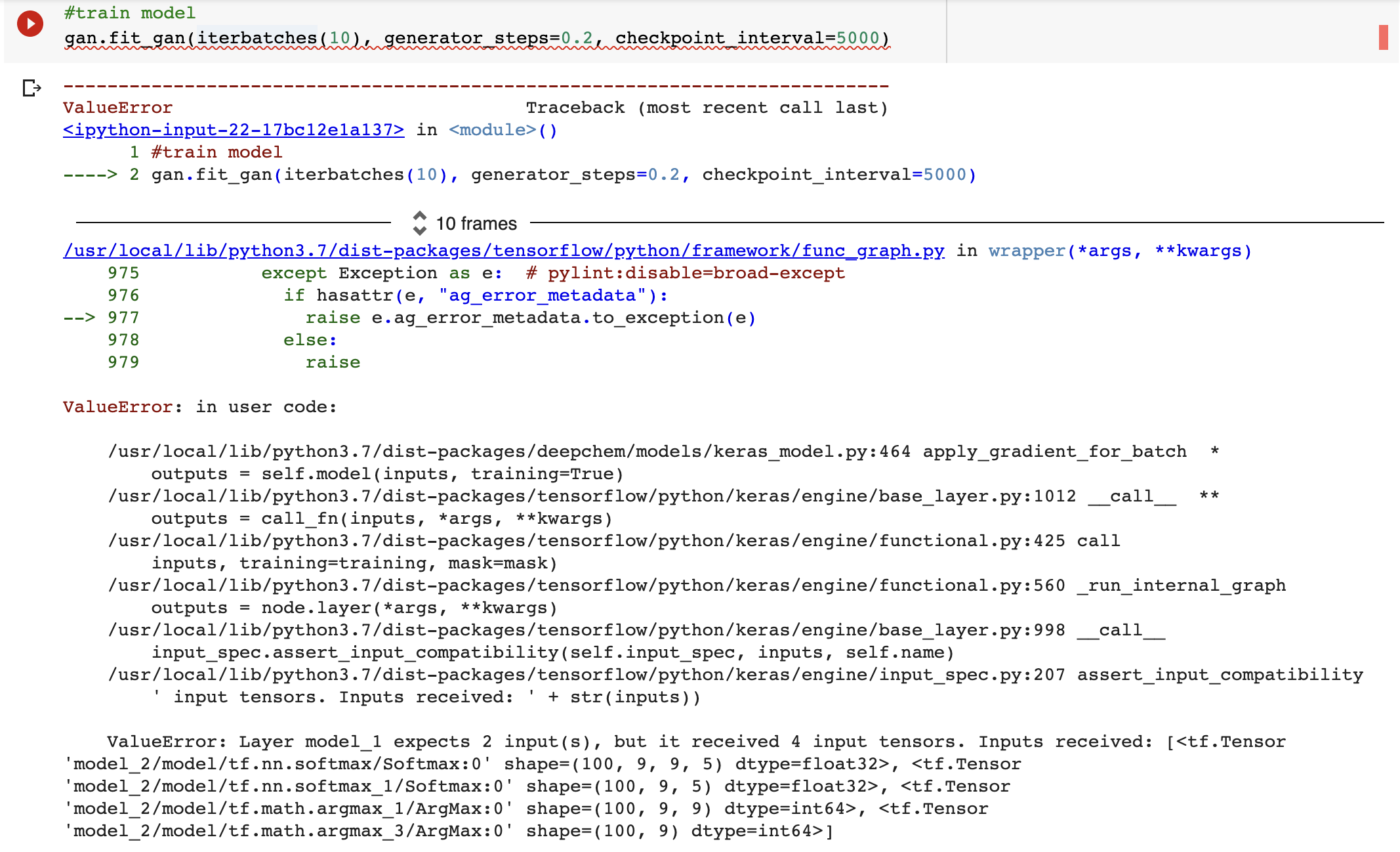
EDIT#2: While going through the tutorial I also received this error while training the model:
> #train model
> gan.fit_gan(iterbatches(10), generator_steps=0.2, checkpoint_interval=5000)
ValueError: Layer model_1 expects 2 input(s), but it received 4 input tensors. Inputs received: [<tf.Tensor ‘model_2/model/tf.nn.softmax/Softmax:0’ shape=(100, 9, 9, 5) dtype=float32>, <tf.Tensor ‘model_2/model/tf.nn.softmax_1/Softmax:0’ shape=(100, 9, 5) dtype=float32>, <tf.Tensor ‘model_2/model/tf.math.argmax_1/ArgMax:0’ shape=(100, 9, 9) dtype=int64>, <tf.Tensor ‘model_2/model/tf.math.argmax_3/ArgMax:0’ shape=(100, 9) dtype=int64>]